 A University of Southern California (USC)-led team has now compiled, archived and shared one of the largest open-source data sets of brain scans from stroke patients via a study. The data set, known as Anatomical Tracings of Lesion After Stroke (ATLAS), is now available for download; researchers around the world are already using the scans to develop and test algorithms that can automatically process MRI images from stroke patients.
A University of Southern California (USC)-led team has now compiled, archived and shared one of the largest open-source data sets of brain scans from stroke patients via a study. The data set, known as Anatomical Tracings of Lesion After Stroke (ATLAS), is now available for download; researchers around the world are already using the scans to develop and test algorithms that can automatically process MRI images from stroke patients.
In the long run, scientists hope to identify biological markers that forecast which patients will respond to various rehabilitation therapies and personalise treatment plans accordingly.
Stroke is the leading cause of disability in adults, affecting more than 15m people worldwide each year, according to the World Health Organisation. During a stroke, blood flow to part of the brain is cut off. Without oxygen, brain cells die and cease to function.
The damaged area, known as a lesion, is what researchers and clinicians study as they design, test and implement recovery programs. Typically, neuroanatomy experts manually draw boundaries around the lesions – in a process called segmentation – but researchers hope to automate this practice so they can examine more images.
"One of our goals is to meta-analyse thousands of stroke MRIs from around the world to understand how the lesions impact recovery," said Sook-Lei Liew, lead author of the study and assistant professor with joint appointments at the Mark and Mary Stevens Neuroimaging and Informatics Institute (INI) within the Keck School of Medicine of USC, the Chan division of occupational science and occupational therapy, the division of bio-kinesiology and physical therapy and the USC Viterbi School of Engineering.
"We can't do it by hand at the scale of thousands, so we are really interested in helping find better automated ways, using machine learning and computer vision, to identify the lesions and have machines draw those boundaries."
"Liew's team is making great strides toward improving patient outcomes following stroke," said Provost Professor Arthur Toga, the INI director. "Several other faculty from the institute and across the university have applied their expertise in machine learning, data visualisation, informatics and neuroradiology to deliver a valuable set of open-source MR images."
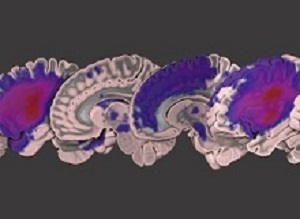
The ATLAS team represents a collaborative effort both within USC and beyond. Dr Hosung Kim, assistant professor of neurology at INI, used a neuroimaging analysis pipeline he developed to help standardize the images in the data set. The institute's Dr Tyler Ard, assistant professor of research, created custom software for advanced visualization of the lesioned data set, rendering it into several extremely high-resolution videos and images. Seventeen other co-authors across the university assisted with analysis, clinical characterisation, and the collection and storage of data.
Data from the project are stored by the INDI, housed at the Child Mind Institute, and by the Inter-University Consortium for Political and Social Research (ICPSR), housed at the University of Michigan. So far, 33 research groups around the world, including from Finland, Iran and Australia, have downloaded the ATLAS data set, which contains 304 manually-segmented MRI scans.
Liew and Kim, along with PhD student Kaori Ito, have already started putting the data set to work. They're testing all of the existing algorithms that attempt to automate the lesion segmentation process to determine which perform the task with greatest accuracy. They presented their work at the annual meeting of the American Society for Neuro-rehabilitation in November and currently have a paper under review.
As predictive algorithms improve, a long-term goal is for clinicians to use MRI to inform decisions about stroke patients' treatment and recovery.
"Ultimately, we would run their data through an automated pipeline that would give us some measures of their likelihood of recovery, or more importantly, their likelihood of responding to different types of therapies," Liew said. "We could then personalise their rehabilitation therapy based on their MRI results and, hopefully, improve their recovery."
Stroke researchers who wish to access the data can download a normalised subset (n=229) from INDI or the full dataset (n=304) from ICPSR.
Abstract
Stroke is the leading cause of adult disability worldwide, with up to two-thirds of individuals experiencing long-term disabilities. Large-scale neuroimaging studies have shown promise in identifying robust biomarkers (e.g., measures of brain structure) of long-term stroke recovery following rehabilitation. However, analyzing large rehabilitation-related datasets is problematic due to barriers in accurate stroke lesion segmentation. Manually-traced lesions are currently the gold standard for lesion segmentation on T1-weighted MRIs, but are labor intensive and require anatomical expertise. While algorithms have been developed to automate this process, the results often lack accuracy. Newer algorithms that employ machine-learning techniques are promising, yet these require large training datasets to optimize performance. Here we present ATLAS (Anatomical Tracings of Lesions After Stroke), an open-source dataset of 304 T1-weighted MRIs with manually segmented lesions and metadata. This large, diverse dataset can be used to train and test lesion segmentation algorithms and provides a standardized dataset for comparing the performance of different segmentation methods. We hope ATLAS release 1.1 will be a useful resource to assess and improve the accuracy of current lesion segmentation methods.
Authors
Sook-Lei Liew, Julia M Anglin, Nick W Banks, Matt Sondag, Kaori L Ito, Hosung Kim, Jennifer Chan, Joyce Ito, Connie Jung, Nima Khoshab, Stephanie Lefebvre, William Nakamura, David Saldana, Allie Schmiesing, Cathy Tran, Danny Vo, Tyler Ard, Panthea Heydari, Bokkyu Kim, Lisa Aziz-Zadeh, Steven C Cramer, Jingchun Liu, Surjo Soekadar, Jan-Egil Nordvik, Lars T Westlye, Junping Wang, Carolee Winstein, Chunshui Yu, Lei Ai, Bonhwang Koo, R Cameron Craddock, Michael Milham, Matthew Lakich, Amy Pienta, Alison Stroud
[link url="http://news.usc.edu/136878/database-brain-images-big-data-to-boost-stroke-recovery/"]University of Southern California material[/link]
[link url="https://www.nature.com/articles/sdata201811"]Scientific Data abstract[/link]
